Chapter 4 Prediction and Bayesian Inference
4.1 Import Data
lottery = read.table("TXTData/Lottery.txt", sep ="\t", quote = "",header=TRUE)
#lottery=read.table(choose.files(), header=TRUE, sep="\t")State of Wisconsin lottery administrators provided weekly lottery sales data. We consider online lottery tickets that are sold by selected retail establishments in Wisconsin. These tickets are generally priced at $1.00, so the number of tickets sold equals the lottery revenue. We analyze lottery sales (OLSALES) over a forty-week period, April, 1998 through January, 1999, from fifty randomly selected ZIP codes within the state of Wisconsin. We also consider the number of retailers within a ZIP code for each time (NRETAIL).
| Variable | Description |
|---|---|
| OLSALES | Online lottery sales to individual consumers |
| NRETAIL | Number of listed retailers |
| PERPERHH | Persons per household MEDSCHYR Median years of schooling |
| MEDHVL | Median home value in $1000s for owner-occupied homes PRCRENT |
| PRC55P | Percent of population that is 55 or older |
| HHMEDAGE | Household median age |
| MEDINC | Estimated median household income, in $1000s |
| POPULATN | Population, in thousands |
#EXTRACT TIME - INVARIANT INFORMATION TO ANALYZE
mzip=d=as.data.frame(t(sapply(split(lottery[, c("NRETAIL", "PERPERHH", "OLSALES", "MEDSCHYR", "MEDHVL", "PRCRENT", "PRC55P", "HHMEDAGE", "MEDINC", "POPULATN")], lottery$ZIP),function(x) colMeans(x))))
# Extract time invariant information to analyze
# Notice: the code for this part on website is wrong.4.2 Example: Forecasting Wisconsin Lottery Sales (Page 138)
In this section, we forecast the sale of state lottery tickets from 50 postal (ZIP) codes inWisconsin. Lottery sales are an important component of state revenues. Accurate forecasting helps in the budget-planning process. A model is useful in assessing the important determinants of lottery sales, and understanding the determinants of lottery sales is useful for improving the design of the lottery sales system. Additional details of this study are in Frees and Miller (2003O).
4.2.1 TABLE 4.2: Time - invariant summary statistics
summary(mzip[,c("NRETAIL", "PERPERHH", "OLSALES", "MEDSCHYR", "MEDHVL", "PRCRENT", "PRC55P", "HHMEDAGE", "MEDINC", "POPULATN")]) ## NRETAIL PERPERHH OLSALES MEDSCHYR
## Min. : 1.000 Min. :2.200 Min. : 189.0 Min. :12.20
## 1st Qu.: 3.000 1st Qu.:2.600 1st Qu.: 821.3 1st Qu.:12.50
## Median : 6.362 Median :2.700 Median : 2426.4 Median :12.60
## Mean :11.942 Mean :2.706 Mean : 6494.8 Mean :12.70
## 3rd Qu.:15.312 3rd Qu.:2.800 3rd Qu.:10016.5 3rd Qu.:12.78
## Max. :68.625 Max. :3.200 Max. :33181.4 Max. :15.90
## MEDHVL PRCRENT PRC55P HHMEDAGE
## Min. : 34.50 Min. : 6.00 Min. :25.0 Min. :41.00
## 1st Qu.: 43.77 1st Qu.:19.25 1st Qu.:35.0 1st Qu.:46.00
## Median : 53.90 Median :24.00 Median :40.0 Median :48.00
## Mean : 57.09 Mean :24.68 Mean :39.7 Mean :48.76
## 3rd Qu.: 66.47 3rd Qu.:27.00 3rd Qu.:44.0 3rd Qu.:51.00
## Max. :120.00 Max. :62.00 Max. :56.0 Max. :59.00
## MEDINC POPULATN
## Min. :27.90 Min. : 0.280
## 1st Qu.:38.17 1st Qu.: 1.964
## Median :43.10 Median : 4.405
## Mean :45.12 Mean : 9.311
## 3rd Qu.:53.62 3rd Qu.:15.446
## Max. :70.70 Max. :39.098# STANDARD DEVIATION
sqrt(diag(var(mzip[,c("NRETAIL", "PERPERHH", "OLSALES", "MEDSCHYR", "MEDHVL", "PRCRENT", "PRC55P", "HHMEDAGE", "MEDINC", "POPULATN")]))) ## NRETAIL PERPERHH OLSALES MEDSCHYR MEDHVL
## 13.2918231 0.2093820 8103.0125037 0.5514212 18.3731152
## PRCRENT PRC55P HHMEDAGE MEDINC POPULATN
## 9.3425513 7.5112161 4.1431527 9.7835616 11.09815704.2.2 FIGURE 4.2: Look at the relationship
Figure 4.2 shows a positive relationship between average online sales and population. Further, the ZIP code corresponding to the city of Kenosha, Wisconsin, has unusually large average sales for its population size.
plot(OLSALES ~ POPULATN, data = mzip, xlab="", ylab="", xaxt="n", yaxt="n",pch="o", las=1, cex=1)
axis(2, at=seq(0, 40000, by=10000), las=1, font=10, cex=0.005, tck=0.01)
axis(2, at=seq(0, 40000, by=1000), lab=F, tck=0.005)
axis(1, at=seq(0,40, by=10), font=10, cex=0.005, tck=0.01)
axis(1, at=seq(0,40, by=1), lab=F, tck=0.005)
mtext("Average Lottery Sales", side=2, line=-3.5, at=36000, font=10, cex=1, las=1)
mtext("Population in Thousands", side=1, line=2, at=20, font=10, cex=1, las=1)
4.2.3 Sorting the data by zip then combine vectors into another data.frame
lottery$logsales<-log10(lottery$OLSALES)
m<-order(lottery$ZIP, lottery$TIME, lottery$OLSALES,lottery$logsales)
index<-as.data.frame(cbind(lottery$ZIP[m],lottery$TIME[m],lottery$OLSALES[m],lottery$logsales[m]))
names(index)<-c("ZIP", "TIME", "OLSALES", "LOGSALES")4.2.4 FIGURE 4.3: Lottery vs. week number
Figure 4.3 presents a multiple time-series plot of (weekly) sales over time. Here, each line traces the sales patterns for a particular ZIP code. This figure shows the dramatic increase in sales for most ZIP codes, at approximately weeks 8 and 18.
plot(OLSALES ~ TIME, data = lottery, axes=F, ylab="", xlab="", xaxt="n", yaxt="n")
for (i in index$ZIP) {
lines(OLSALES ~ TIME, data = subset(index, ZIP == i)) }
axis(1, at=seq(0,40, by=1), labels=F, tck=0.005)
axis(1, at=seq(0,40, by=10), cex=0.005, tck=0.01)
mtext("Week Number", side=1, line=2.5, cex=1, font=10)
axis(2, at=seq(0, 300000, by=10000), labels=F, tck=0.005)
axis(2, at=seq(0, 305000, by=100000), las=1, cex=0.005, tck=0.01)
mtext("Lottery Sales", side=2, line=-3, at=310000, font=10, cex=1, las=1)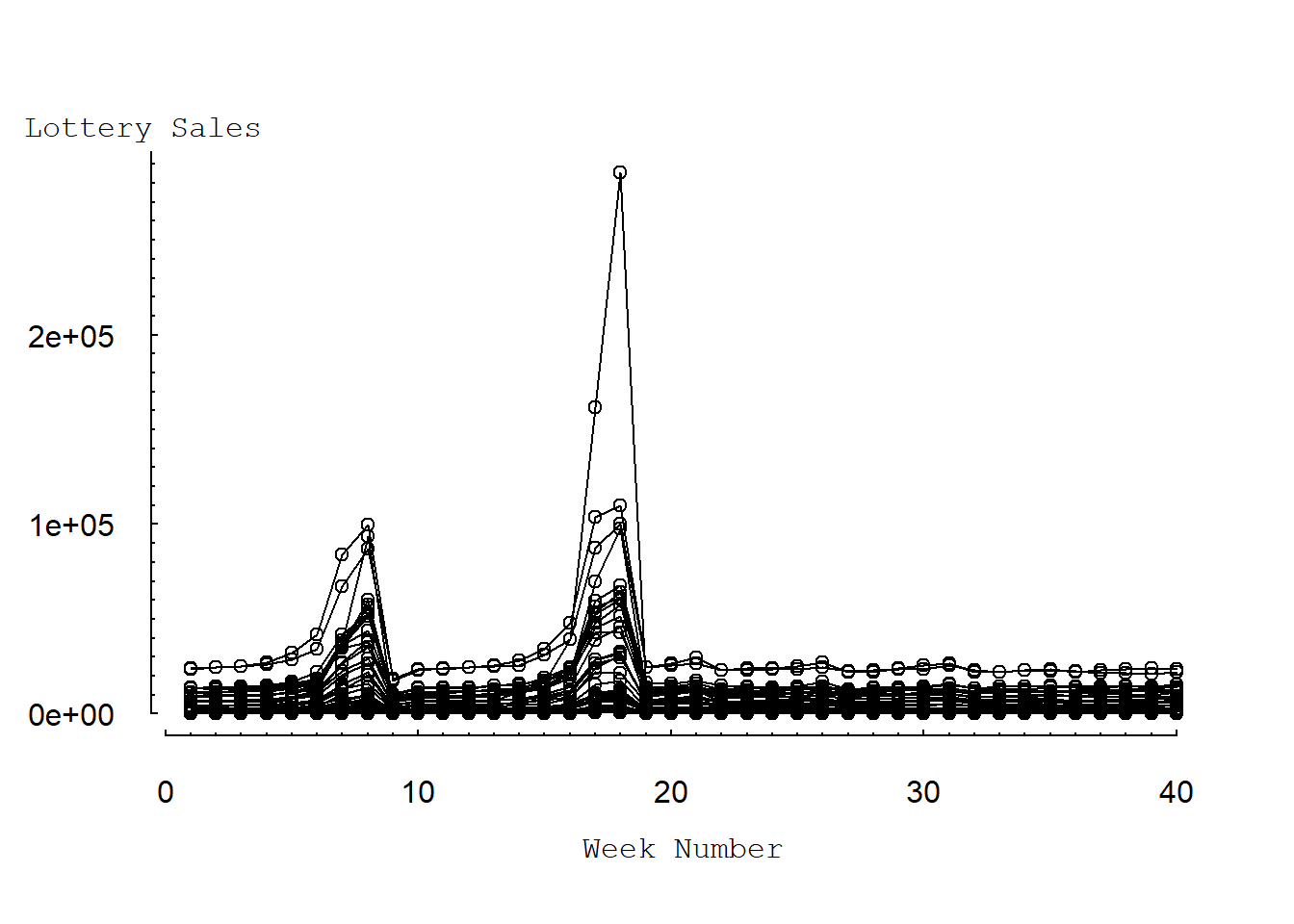
Another way of producing multiple time series graph by using trellis xyplot:
library(lattice)
trellis.device(color=F) # telling the trellis device to mimic 'black and white'
xyplot(OLSALES ~ TIME, data=index, groups=ZIP, scales=list(y=list(at=seq(0, 300000,100000), tck=.01)), panel=panel.superpose, pch=16, lty=1, type="b")
#ChECK LOG VALUES
lottery$logsales<-log10(lottery$OLSALES)
lottery$lnsales<-log(lottery$OLSALES)4.2.5 FIGURE 4.4: Log lottery vs week number
Figure 4.4 shows the same information as in Figure 4.3 but on a common (base 10) logarithmic scale. Here, we still see the effects of the PowerBall jackpots on sales. However, Figure 4.4 suggests a dynamic pattern that is common to all ZIP codes. Specifically, logarithmic sales for each ZIP code are relatively stable with the same approximate level of variability. Further, logarithmic sales for each ZIP code peak at the same time, corresponding to large PowerBall jackpots.
#FIGURE 4.4 LOG LOTTERY vs WEEK NUMBER
plot(LOGSALES ~ TIME, data = index, type="p", axes=F, ylab="", xlab="", pch=16, mkh=0.0001, lwd=0.5)
axis(1, at=seq(0,40, by=1), labels=F, tck=0.005)
axis(1, at=seq(0,40, by=10), cex=0.4, tck=0.01)
mtext("Week Number", side=1, line=2.5, cex=0.7, font=10)
axis(2, at=seq(0, 6, by=0.1), labels=F, tck=0.005)
axis(2, at=seq(0, 6, by=1), las=1, cex=0.4, tck=0.01)
mtext("Logarithmic Lottery Sales", side=2, line=-1, at=5.8, font=10, cex=0.7, las=1)
for (i in index$ZIP) {
lines(LOGSALES ~ TIME, data=subset(index, ZIP==i)) }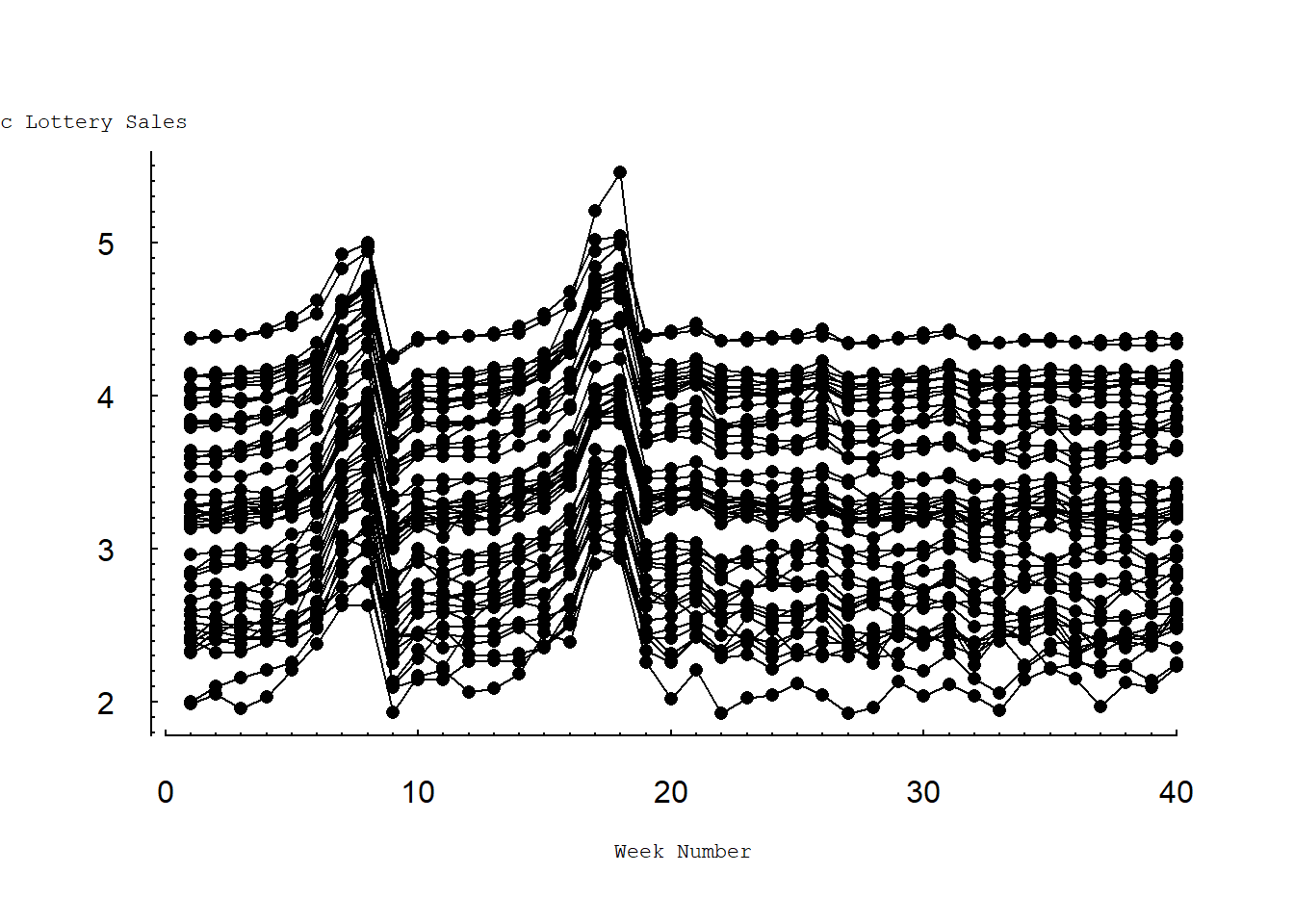
4.3 Create model development sample
Lottery=lottery
Lottery$LNSALES<-log(Lottery$OLSALES)
Lottery2<-subset(Lottery, Lottery$TIME<36)4.3.1 MODEL 1. Pooled cross-setional model
lm1<-lm(LNSALES~PERPERHH+MEDSCHYR+MEDHVL+PRCRENT+PRC55P+HHMEDAGE+MEDINC+POPULATN+NRETAIL, data=Lottery2)
summary(lm1)#Pooled cross-sectional model ##
## Call:
## lm(formula = LNSALES ~ PERPERHH + MEDSCHYR + MEDHVL + PRCRENT +
## PRC55P + HHMEDAGE + MEDINC + POPULATN + NRETAIL, data = Lottery2)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.9743 -0.6012 -0.0774 0.5430 4.2015
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 13.821060 1.339594 10.317 < 2e-16 ***
## PERPERHH -1.084705 0.160224 -6.770 1.76e-11 ***
## MEDSCHYR -0.821644 0.069049 -11.899 < 2e-16 ***
## MEDHVL 0.013822 0.002662 5.192 2.33e-07 ***
## PRCRENT 0.031820 0.003738 8.512 < 2e-16 ***
## PRC55P -0.069578 0.013397 -5.194 2.30e-07 ***
## HHMEDAGE 0.118136 0.020961 5.636 2.03e-08 ***
## MEDINC 0.043373 0.005304 8.177 5.53e-16 ***
## POPULATN 0.057025 0.006060 9.410 < 2e-16 ***
## NRETAIL 0.021278 0.004076 5.220 2.00e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.8365 on 1740 degrees of freedom
## Multiple R-squared: 0.6963, Adjusted R-squared: 0.6947
## F-statistic: 443.3 on 9 and 1740 DF, p-value: < 2.2e-164.3.2 MODEL 2. Error components model
library(nlme)
lme1<-lme(LNSALES~PERPERHH+MEDSCHYR+MEDHVL+PRCRENT+PRC55P+HHMEDAGE+MEDINC+POPULATN+NRETAIL, data=Lottery2, random=~1|ZIP, method="REML")
# NOTE* THE DEFAULT METHOD IN lme IS "REML"
# Use REML method in estimating fixed effects beta coefficients
summary(lme1)## Linear mixed-effects model fit by REML
## Data: Lottery2
## AIC BIC logLik
## 2907.889 2973.428 -1441.944
##
## Random effects:
## Formula: ~1 | ZIP
## (Intercept) Residual
## StdDev: 0.77897 0.5130729
##
## Fixed effects: LNSALES ~ PERPERHH + MEDSCHYR + MEDHVL + PRCRENT + PRC55P + HHMEDAGE + MEDINC + POPULATN + NRETAIL
## Value Std.Error DF t-value p-value
## (Intercept) 18.095695 7.316764 1699 2.473183 0.0135
## PERPERHH -1.287021 0.886172 41 -1.452337 0.1540
## MEDSCHYR -1.077937 0.375131 41 -2.873491 0.0064
## MEDHVL 0.007360 0.014633 41 0.502935 0.6177
## PRCRENT 0.026321 0.020660 41 1.274032 0.2098
## PRC55P -0.072547 0.074259 41 -0.976939 0.3343
## HHMEDAGE 0.118637 0.116199 41 1.020986 0.3132
## MEDINC 0.045540 0.029396 41 1.549194 0.1290
## POPULATN 0.121851 0.027529 41 4.426231 0.0001
## NRETAIL -0.027177 0.017420 1699 -1.560055 0.1189
## Correlation:
## (Intr) PERPER MEDSCH MEDHVL PRCREN PRC55P HHMEDA MEDINC POPULA
## PERPERHH -0.632
## MEDSCHYR -0.745 0.204
## MEDHVL 0.303 0.093 -0.394
## PRCRENT -0.198 0.402 -0.258 0.008
## PRC55P 0.146 0.236 -0.018 0.069 0.039
## HHMEDAGE -0.461 0.049 0.109 -0.128 0.151 -0.898
## MEDINC -0.171 -0.013 0.080 -0.653 0.214 0.392 -0.200
## POPULATN 0.180 -0.021 -0.228 -0.171 -0.287 -0.035 0.035 -0.050
## NRETAIL -0.210 0.082 0.246 0.159 0.096 0.014 -0.002 -0.027 -0.847
##
## Standardized Within-Group Residuals:
## Min Q1 Med Q3 Max
## -2.06921597 -0.49881173 -0.29717361 0.02767368 6.60150830
##
## Number of Observations: 1750
## Number of Groups: 50# CHECK AUTOCORRELATION PATTERNS
ACF(lme1, maxlag=10) #Obtain ACF of residuals from lme1## lag ACF
## 1 0 1.00000000
## 2 1 0.52724776
## 3 2 0.10000857
## 4 3 -0.03788895
## 5 4 -0.15969734
## 6 5 -0.23410399
## 7 6 -0.24984691
## 8 7 -0.18355756
## 9 8 -0.02825433
## 10 9 0.19456638
## 11 10 0.47143962
## 12 11 0.17601528
## 13 12 -0.10862546
## 14 13 -0.16449717
## 15 14 -0.31225995
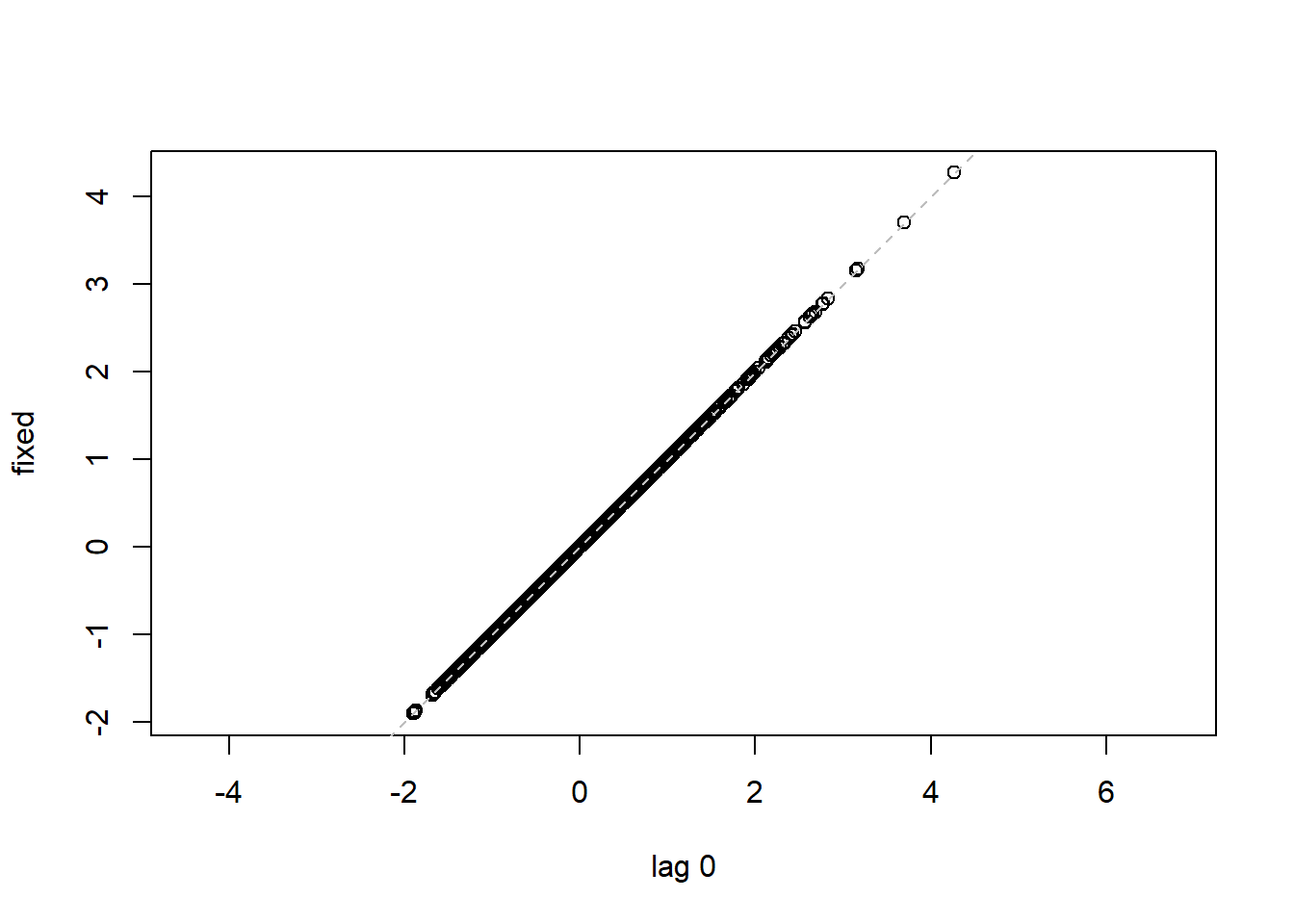
## 16 15 -0.40819498lag.plot(lme1$residuals, lags=-1) #Autocorrelation patterns one lag, needs to refine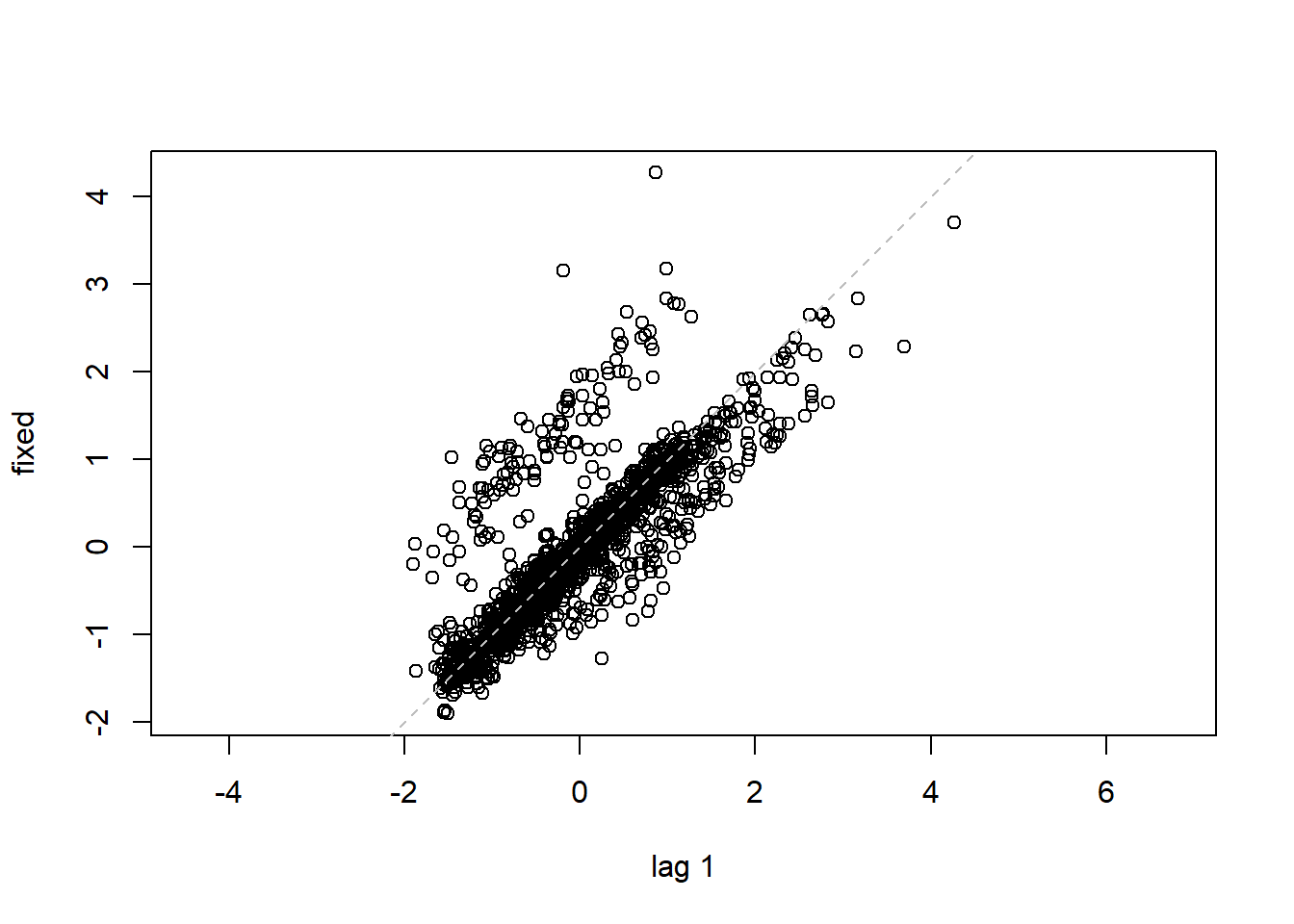
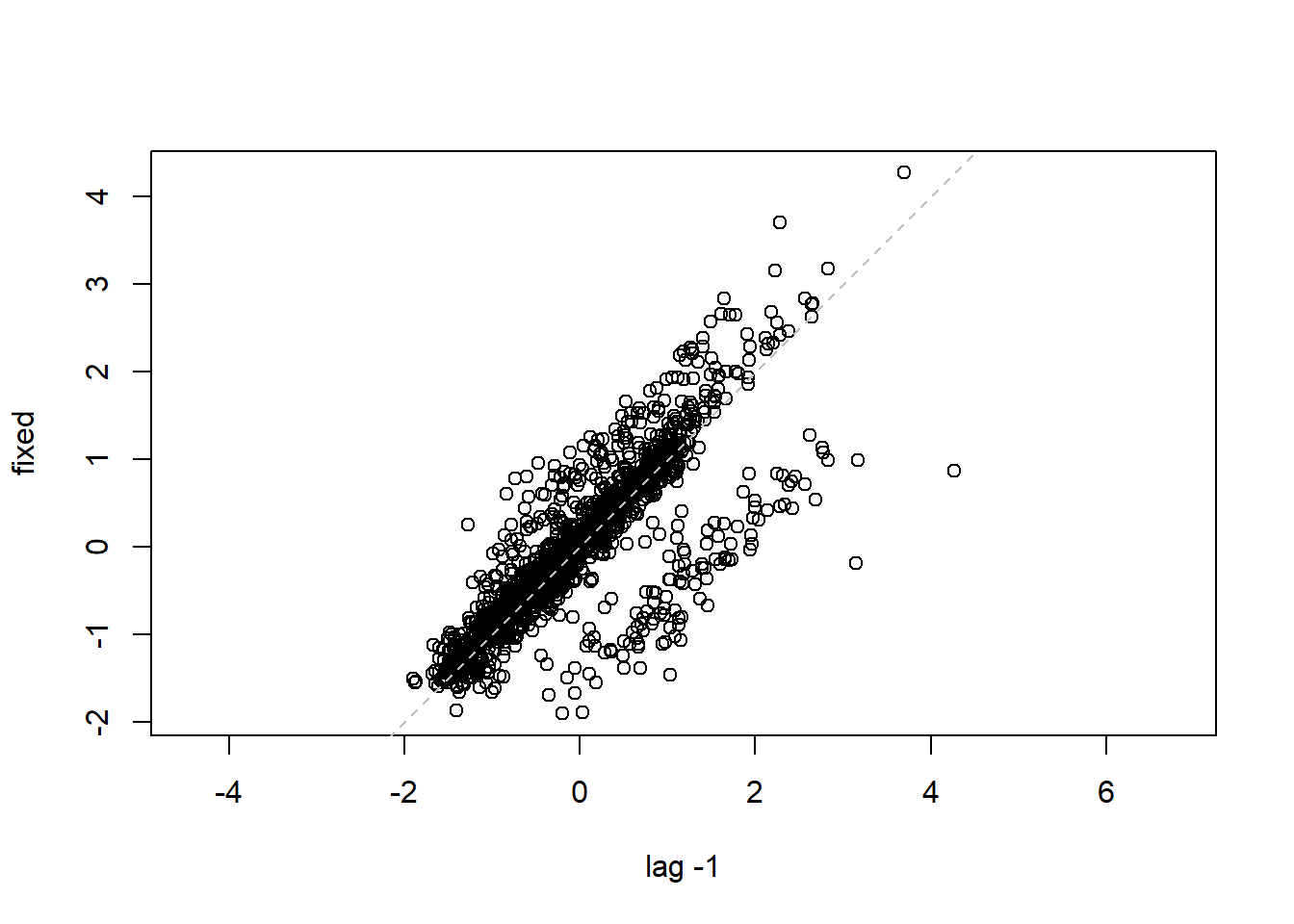
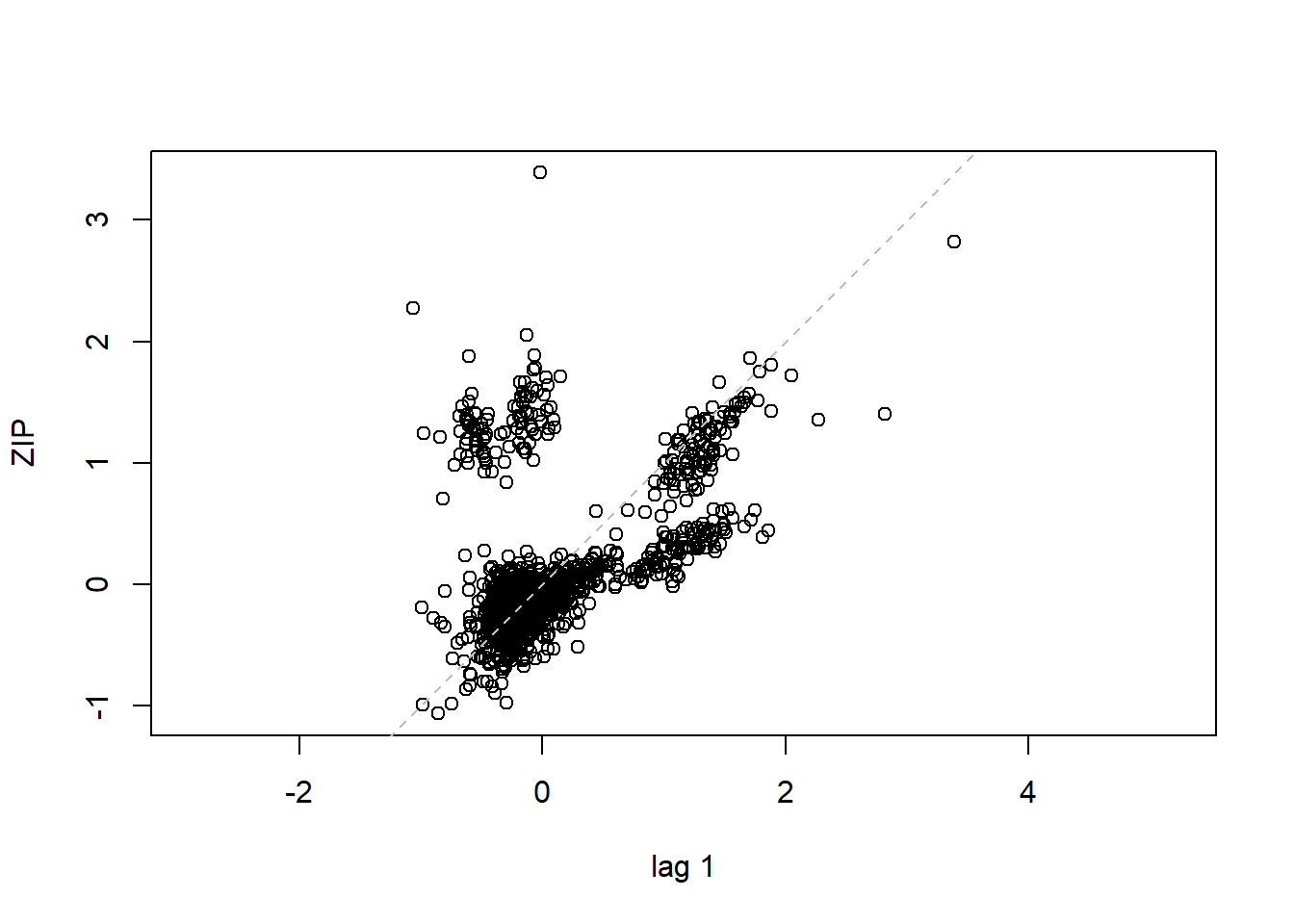
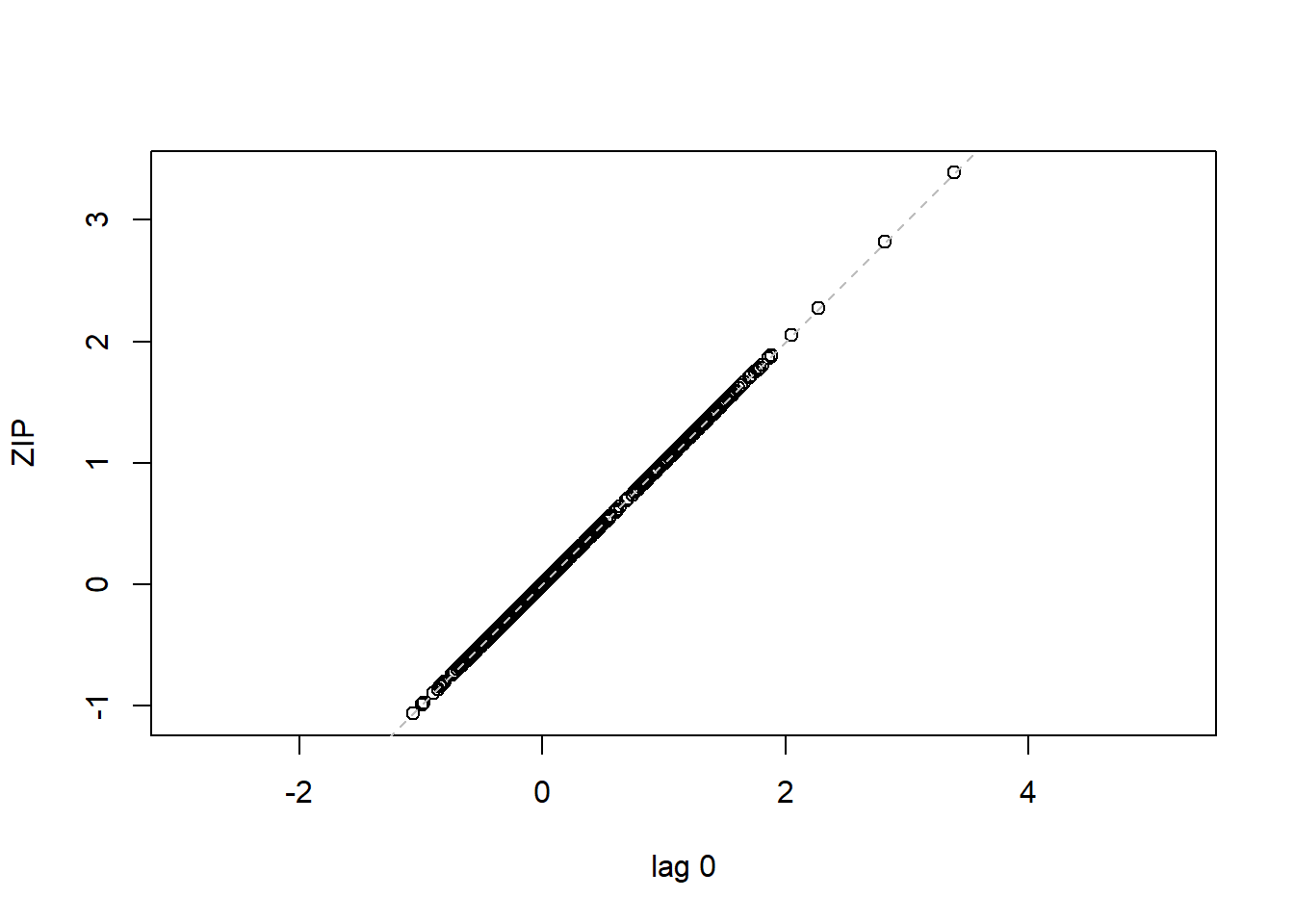
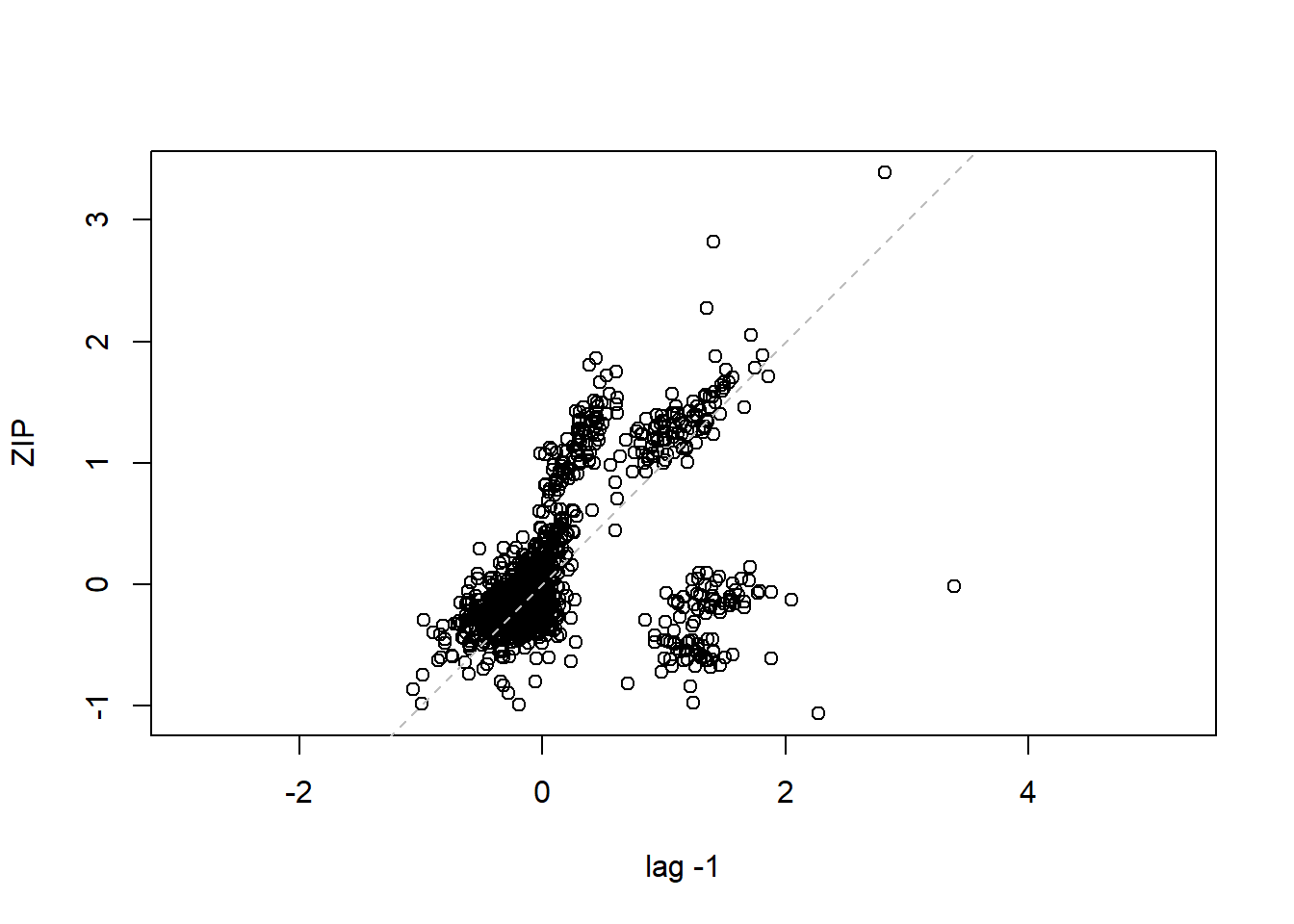
4.3.4 MODEL 4. More parsimonious random effects model
With fewer covariates:
lme3<-lme(LNSALES~MEDSCHYR+POPULATN, data=Lottery2, random=~1|ZIP, correlation=corAR1(form=~TIME|ZIP))#The pooled cross-sectional model with autocorrelated errors
#Default method for gls is reml, gls can be viewed as an lme function without the argument random
library(nlme)
gls1<-gls(LNSALES~PERPERHH+MEDSCHYR+MEDHVL+PRCRENT+PRC55P+HHMEDAGE+MEDINC+POPULATN+NRETAIL, data=Lottery2, correlation=corAR1(form=~TIME|ZIP))
summary(gls1)## Generalized least squares fit by REML
## Model: LNSALES ~ PERPERHH + MEDSCHYR + MEDHVL + PRCRENT + PRC55P + HHMEDAGE + MEDINC + POPULATN + NRETAIL
## Data: Lottery2
## AIC BIC logLik
## 2505.647 2571.186 -1240.823
##
## Correlation Structure: AR(1)
## Formula: ~TIME | ZIP
## Parameter estimate(s):
## Phi
## 0.8240088
##
## Coefficients:
## Value Std.Error t-value p-value
## (Intercept) 13.596133 3.858304 3.523863 0.0004
## PERPERHH -1.063225 0.461639 -2.303151 0.0214
## MEDSCHYR -0.820030 0.198788 -4.125146 0.0000
## MEDHVL 0.012934 0.007667 1.686984 0.0918
## PRCRENT 0.032524 0.010771 3.019630 0.0026
## PRC55P -0.072183 0.038596 -1.870207 0.0616
## HHMEDAGE 0.122852 0.060391 2.034284 0.0421
## MEDINC 0.043223 0.015281 2.828636 0.0047
## POPULATN 0.058989 0.017339 3.402168 0.0007
## NRETAIL 0.020143 0.011647 1.729449 0.0839
##
## Correlation:
## (Intr) PERPER MEDSCH MEDHVL PRCREN PRC55P HHMEDA MEDINC POPULA
## PERPERHH -0.633
## MEDSCHYR -0.754 0.213
## MEDHVL 0.275 0.101 -0.359
## PRCRENT -0.208 0.405 -0.237 0.018
## PRC55P 0.142 0.237 -0.016 0.069 0.039
## HHMEDAGE -0.454 0.049 0.107 -0.127 0.151 -0.898
## MEDINC -0.165 -0.014 0.075 -0.651 0.211 0.392 -0.200
## POPULATN 0.242 -0.056 -0.295 -0.212 -0.280 -0.035 0.029 -0.030
## NRETAIL -0.267 0.107 0.310 0.202 0.124 0.018 -0.001 -0.033 -0.898
##
## Standardized residuals:
## Min Q1 Med Q3 Max
## -2.26140351 -0.64050416 -0.02884818 0.69976712 5.08212114
##
## Residual standard error: 0.8427768
## Degrees of freedom: 1750 total; 1740 residual#getVarCov(gls1)
anova(gls1,lm1)## Model df AIC BIC logLik Test L.Ratio p-value
## gls1 1 12 2505.647 2571.186 -1240.823
## lm1 2 11 4430.126 4490.204 -2204.063 1 vs 2 1926.479 <.0001#using anova for the models with serial correlation and the pooled cross–sectional regression.